DILIsym is Quantitative Systems Toxicology (QST) software designed to be used during drug development to provide an indication of the potential drug-induced liver injury (DILI) hazard posed by individual molecules and/or to provide deeper insight into the mechanisms responsible for observed DILI responses at various stages of the development process.

-
DILIsym application examples include
Important DILIsym application examples include:

- Comparison of ubrogepant with prior in-class molecules for DILI potential, which was included in an FDA review of the ubrogepant safety data
- Comparison of lixivaptan’s liver safety profile with tolvaptan, which was submitted to the FDA as part of the new IND application for lixivaptan
- Mechanistic analysis of acetaminophen’s carcinogenic potential, which was submitted to the California Office of Environmental Health Hazard Assessment (OEHHA) regarding Proposition 65 as part of the weight of evidence suggesting no cancer risk; FDA agreed
- Mechanistic analyses of the liver safety of remdesivir, tolvaptan, pexidartinib, and TAK-875
- Comparison of the liver safety mechanisms of several macrolide antibiotics as part of the solithromycin new drug application advisory committee meeting
- Liver safety biomarker analyses for entolimod and GGF2
- Dose optimization for a novel antibiotic, BAL30072
-
DILIsym integrations
DILIsym simulations integrate compound exposure with compound mediated mechanisms of intrinsic drug toxicity, and with inter-individual variability. The intersection of these is prediction of hepatotoxicity for:
- Phase II, III, or IV clinical trials in patients
- First-in-human clinical trials in healthy volunteers
- Small preclinical species
- Large preclinical species
DILIsym is a “middle-out”, multi‑scale representation of drug-induced liver injury. It includes key liver cell populations (e.g., hepatocytes, Kupffer cells), bile duct cells (cholangiocytes), intracellular biochemical systems (e.g., mitochondrial function), and whole body dynamics (e.g., drug distribution and metabolism). DILIsym represents these biochemical and physiological processes for mice, rats, dogs, and humans, based on a substantial amount of publicly-available literature. Through the generation of SimPops® (a collection of simulated patients forming a simulated population), DILIsym also includes inter‑individual variability.
DILIsym includes a graphical user interface (GUI) to facilitate in silico investigations. The GUI permits the specification of new simulated experiments, including compound, dose, dosing frequency, duration, mechanism(s), and species. Simulations can be performed on individuals or populations. Simulation results can be visualized within the GUI or exported for further analysis in other software. Simulations can also be run with the command-line version of DILIsym, making integration into a stream-lined workflow possible. A high performance grid license (HPGL) option is available via licensing (or included with DILI-sim Initiative membership), which allows for use of an unlimited number of command-line-based simulation engines and enables users to scale up and complete large batches of simulations on internal or external compute clusters.
DILIsym prospectively supports key management decisions by providing information about potential drug-induced liver injury risk in upcoming experimental or clinical study designs, as well as mechanistic rationale for the underlying biochemical events that could cause liver toxicity. DILIsym also supports key management decisions about compounds for which liver transaminase elevations have already been observed by identifying patient screening or dosing protocols to mitigate risk. Additionally, DILIsym can mechanistically interpret existing data on liver transaminase elevations to predict the underlying level of hepatocyte loss and rate of liver recovery, supporting the distinction of innocuous vs. dangerous liver signals.
-
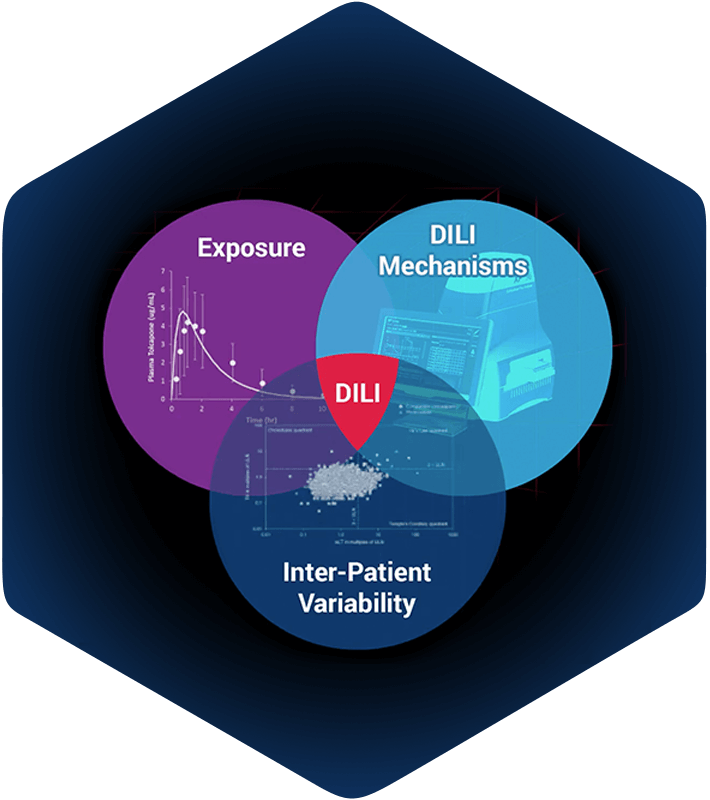
Conceptual framework for DILIsym development
DILIsym has been developed within a conceptual framework (Figure 1), wherein liver compound exposure (parent and/or metabolite) can engage various mechanisms of intrinsic toxicity. The mechanisms currently included are: (1) inhibition of bile acid transporters leading to disruptions in bile acid homeostasis, (2) alteration of mitochondrial function, (3) generation of oxidative stress, and (4) cholestatic injury to bile duct cells via disruption in the ratio between phospholipids and bile acids in the bile. Alone or in combination, these mechanisms can lead to hepatocyte cell death via apoptosis or necrosis. Hepatocyte necrosis releases damage associated molecular pattern (DAMP) molecules, capable of activating innate immune cells (e.g., resident Kupffer cells). Activated immune cells produce mediators that may serve to amplify the ongoing injury and/or may promote liver regeneration. Immune cells may also accumulate due to ongoing inflammatory processes. Importantly, while the various elements described thus far can be sufficient to drive liver injury, they may also provide the setting in which an adaptive immune response can develop. Adaptive immune responses are not currently included in DILIsym but are part of ongoing development to be included in future versions of the modeling software.
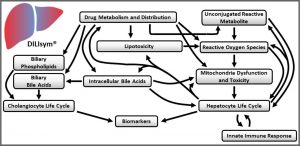
Figure 1. Conceptual framework for DILIsym development. -
What DILIsym includes?
DILIsym includes multiple sub-models which are solved in an integrated fashion to generate simulation results. Sub-models include intracellular biochemical systems that may be impacted by compound exposure (e.g., bile acid homeostasis, mitochondrial function, oxidative stress), the hepatocyte and cholangiocyte life cycles, and stimulation of innate immune responses. To facilitate comparison with high-level clinical and preclinical readouts, DILIsym includes the release of biomarkers (e.g., ALT, AST, ALP) from hepatocytes in the event of cell death, as well as increases in total bilirubin, reflecting compromised liver function due to hepatocyte loss or direct bilirubin-processing enzyme/transporter compound effects. DILIsym also includes emerging biomarkers (e.g., GLDH, miR-122, CK-18) that may provide additional mechanistic information. More detailed descriptions of the sub-models are provided in the links below. Specific data used in design and development are available to licensees through the DILIsym modeling software documentation.
Finally, DILIsym includes SimPops, comprising simulated individuals expressing variability in key parameters related to susceptibility to various mechanisms of toxicity. SimPops permit the researcher to consider the effect of inter-individual variability on drug toxicity.
DILIsym Submodules
DILIsym is Quantitative Systems Toxicology (QST) software designed to be used during drug development to provide an indication of the potential drug-induced liver injury (DILI) hazard posed by individual molecules and/or to provide deeper insight into the mechanisms responsible for observed DILI responses at various stages of the development process.
DILIsym development is supported by a pre-competitive consortium, the DILI-sim Initiative, that includes DILIsym Services and multiple pharmaceutical companies. Member companies have a seat at the table in directing development efforts. The DILIsym development team receives regular feedback from member companies and from the Scientific Advisory Board, which includes key opinion leaders within the field of drug-induced liver injury.
- What are we providing with DILIsym?
- How is DILIsym being applied?
- What's new in DILIsym version X (DSX)?
DILIsym modeling software functions include:
- General population samples: The DILIsym software can be used to test compounds in simulated populations, termed SimPops. Simulated individuals within the SimPops express a wide‑range of response to the various exemplar compounds and can be characterized by extensive variability in their underlying biochemistry.
- DILIsym humans: A middle-out multi-scale representation of human physiology for assessing potential drug-induced liver injury (DILI) hazard in patients. Compound pharmacokinetic (PK) and pharmacodynamics (PD) information can be integrated to predict time profiles of liver enzymes (i.e., alanine aminotransferase, aspartate aminotransferase) and other clinical variables (e.g., bilirubin, prothrombin time, INR), as well as tissue properties (e.g., liver mass, GSH content). Alternate hypotheses regarding the downstream mechanisms of drug action can be investigated, including increased reactive oxygen/nitrogen species, ATP utilization, direct hepatocyte necrosis, and inhibition of bile acid transporters.
- DILIsym dogs: A middle-out multi-scale representation of Beagle dog physiology for assessing potential DILI hazard in dogs. Data from Beagle dogs were prioritized among the available datasets to maximize consistency in the representation.
- DILIsym rats: A middle-out multi-scale representation of Sprague-Dawley rat physiology for assessing potential DILI hazard in rats. Data from Sprague-Dawley rats were prioritized among the available datasets to maximize consistency in the representation. Compound pharmacokinetic and pharmacodynamics information can be integrated to predict time profiles as described above.
- DILIsym mice: A middle-out multi-scale representation of C57Bl/6 mouse physiology for assessing potential DILI hazard in mice. Data from C57Bl/6 mice were prioritized among the available datasets to maximize consistency in the representation. Compound pharmacokinetic and pharmacodynamics information can be integrated to predict time profiles as described above.
- Translational research: The ability to integrate your in vitro, small animal, and large animal compound data into a single platform facilitates translational research to better inform program advancement decisions, including experimental design, analyte selection, and timing of sampling.
DILIsym® is computer software, namely, computer software for modeling liver response to a drug or chemical; computer software for conducting simulations of drug-liver interactions; computer software for predicting pharmacokinetic, pharmacodynamic, and liver responses to a drug or chemical.
Key features of the DILIsym modeling software
- In vitro extrapolation to in vivo predictions (IVIVE) of drug-induced liver injury (DILI) hazard in multiple preclinical species and in humans
- Predicted DILI hazard can be traced back to its mechanistic source(s)
- Incorporates inter-individual physiological variability
- Incorporates PK/PD variability
- User-friendly GUI to specify in silico experiments and visualize results
- Transparent design and parameterization
- Inclusive incorporation of the available literature for mechanistic representation of the physiology
- Regularly updated to include leading edge science
- Developed and supported by scientific and technical teams
- Consortium members guide the development of DILIsym and may share data to support model development and improve the collective understanding of DILI
- Informs drug development decisions and risk mitigation strategies
Key benefits of the DILIsym modeling software
- Potential DILI hazard posed by specific molecules
- Impact of alternate pharmacology or experimental protocols on potential DILI hazard
- Mechanisms contributing to potential DILI hazard
- Mechanistic differences in cross-species sensitivity
- Identifies non-standard mechanistically-relevant safety biomarkers of DILI hazard
- Facilitates maximal use of data by integrating nonclinical and clinical data in a single platform
- Allows SimPops customization to reflect clinical population characteristics
- Rapidly realizes implications of “what if” scenarios
- User-friendly for scientists
- Differentiates lead candidates according to DILI potential
DSX is our newest, most exciting release of DILIsym yet! A sample of the enhancements include:
- A complete software redesign that includes command line and graphical interface options and server/cloud computing capability (HPGL)
- NO RELIANCE on MATLAB base or runtime
- 4 NEW exemplar compounds included with varying clinical presentations:
- PF-04895162 (Generaux 2019)
- Efavirenz
- Anastrozole
- Tamoxifen
- 2 NEW SimCohorts that include variability in susceptibility to liver injury and biomarker-related parameters (ALT and bilirubin)