ADMET Predictor’s PCB Module contains models for physicochemical property prediction. Each model was built by Simulations Plus scientists using artificial neural network ensemble (ANNE) technology. The data sets were highly curated in order to generate highly accurate models. The data for our pKa model (S+pKa) was recently expanded in a collaboration with Bayer scientists. This resulted in more accurate predictions for a set of 16,000 compounds that were not used to train the model. Our logP (S+logP) and aqueous solubility (S+Sw) models have been ranked number one in peer reviewed journal articles.1,2 The models in the PCB module are also available in the ADMET Predictor module of GastroPlus® in order to perform PBPK simulations using in silico predicted properties.
01. Discovery
What is the PCB Module?
02. Explore
Physicochemical property prediction
The image below lists the models in ADMET Predictor’s PCB Module.
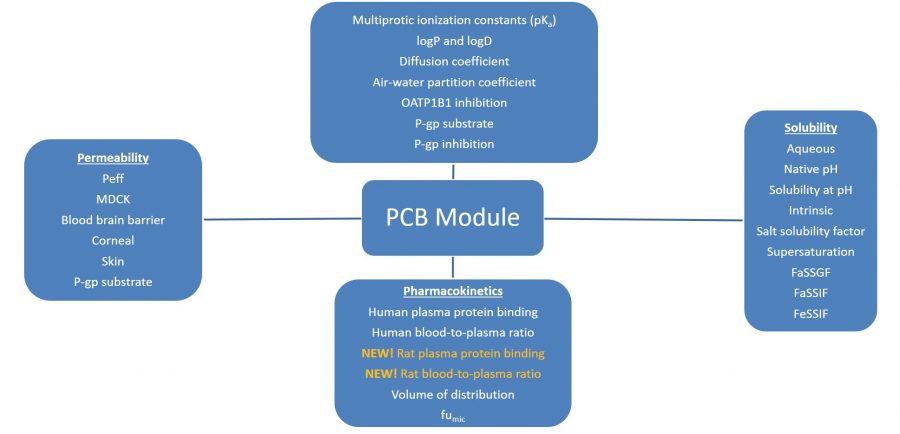
-
New Models created for ADMET Predictor version 12!
Key enhancements include:
- Enhanced Models: New and expanded models offer greater predictive accuracy, with an average 30% increase in training set sizes, for microsome and hepatocyte clearance, protein binding, biorelevant solubilities, MDCK-LE/PAMPA permeability, and more.
- High-Throughput Pharmacokinetics (HTPK): New options for solution dosing, adjusted free fraction outputs, and species-specific simulations enhance the flexibility and precision of HTPK studies.
- Artificial Intelligence-Driven Drug Design (AIDD): Integration of 3D shape matching and tissue sensitivities (based on tissue Kp values) as new objectives, facilitating innovative lead optimization processes.
- New DILI Module: Introduction of the first drug-induced liver injury (DILI) endpoint models to support high-throughput (HT) DILIsym® predictions in early drug development.
- Boosted ANN regression models and 37 new descriptors in ADMET Modeler™
- + more
-
pKa, partition, and transporters models
- Multiprotic pKa model (S+Acidic_pKa, S+Mixed_pKa, S+Basic_pKa) – a thermodynamically accurate multiprotic model for multiple ionization sites based on atomic descriptors and neural networks – not a database lookup!
- logP (S+logP, MlogP) – log of the octanol to water partition coefficient. There are two models, an artificial neural network ensemble (S+logP) and Moriguchi (MlogP)
- logD (S+logD) – estimation of octanol-water distribution coefficient at user-defined pH
- Air-water partition (logHLC) – estimation of air-water partition coefficient (Henry’s Law constant) from US EPA data
- Inhibition of the hepatic OATP1B1 transporter in human (OATP1B1_Inh)
- Inhibition of the intestinal P-gp transporter in human (Pgp_Inh)
- Likelihood of intestinal efflux by P-gp transporter in human (Pgp_Substr)
-
Permeability models
- Human effective permeability (S+Peff) – jejunal Peff
- MDCK apparent permeability (S+MDCK) – in vitro Papp
- Corneal permeability (Perm_Cornea) – ocular permeability through rabbit cornea based on literature data obtained in vitro
- Skin permeability (Perm_Skin) – permeability through human skin of compounds dissolved in aqueous solution; based on literature data
- Blood-brain barrier permeation – there are two models, classification (BBB_Filter) and regression (LogBB). The first classification model has a low cutoff such that compounds that are predicted to have low permeability have very little chance of penetrating the blood-brain barrier. Compounds that are predicted to have high blood-brain barrier penetration should be evaluated with the regression model that predicts the blood to brain concentration ratio.
-
Solubility models
- Aqueous solubility:
- Native solubility (S+Sw) – solubility in pure water
- Native pH at saturation in pure water (S+pH_Satd)
- Intrinsic solubility in pure water (S+S_Intrins)
- Salt solubility factor (SolFactor)
- Water solubility at user-specified pH (S+S_pH)
- Solubility in the simulated gastrointestinal fluids (the models were built using data in our Biorelevant Solubility Database)
- Fasted state simulated gastric fluid solubility (S+FaSSGF)
- Fasted state simulated intestinal fluid (S+FaSSIF)
- Fed state simulated intestinal fluid (S+FeSSIF)
- Supersaturation ratio (SupSatn) – a tendency to supersaturate in water
- Aqueous solubility:
-
Pharmacokinetic models
- Human and rat plasma protein binding as percent unbound (hum_fup% and rat_fup%)
- Human volume of distribution (Vd)
- Human and rat blood-to-plasma concentration ratio (RBP and RBP_rat)
- Fraction unbound in human liver microsomes (S+fumic)
03. Resources
Explore Resources

