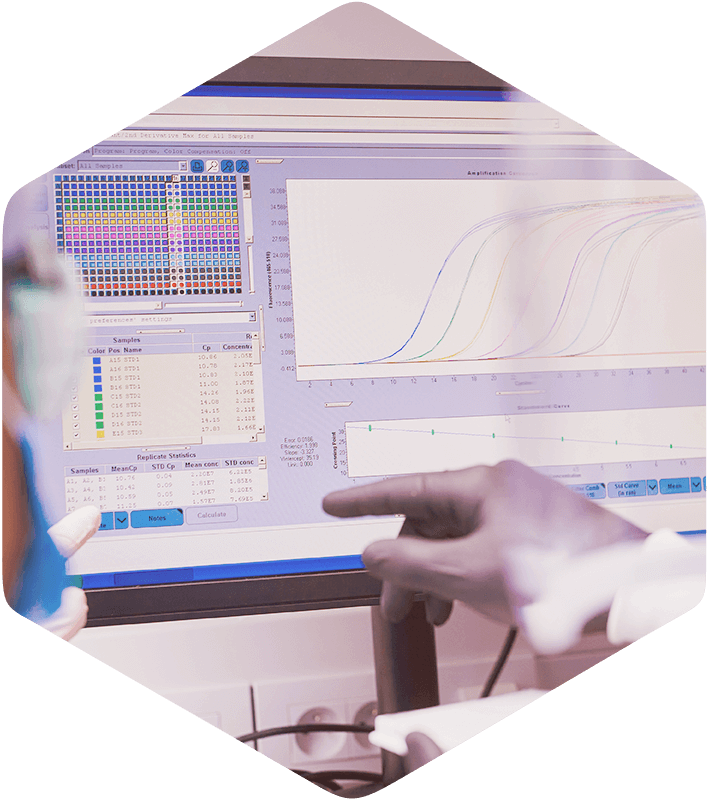
Welcome to DDDPlus™, the industry’s leading mechanistic in vitro dissolution software for formulation and analytical scientists. With DDDPlus, you can model and simulate the in vitro dissolution of active pharmaceutical ingredients (API) and formulation excipients under various experimental conditions in seconds, and begin making informed decisions to help improve your chances for success.

Today, dissolution studies are the most frequently used tools in the development, characterization, and utilization process of pharmaceutical dosage forms, both immediate and controlled-release. As a formulation or CMC scientist, you’re often tasked with designing a product which achieves a target in vitro dissolution rate that will, hopefully, lead to desired in vivo exposure levels. Or, the lifecycle management team wants to assess the possibility of creating a once-a-day formulation which is bioequivalent to a B.I.D. (twice a day) or T.I.D. (three times a day) dosing regimen. You don’t have a lot of time or material available, so you begin manufacturing small lots of products to test under different conditions, hoping something works. What if there was a more efficient strategy?
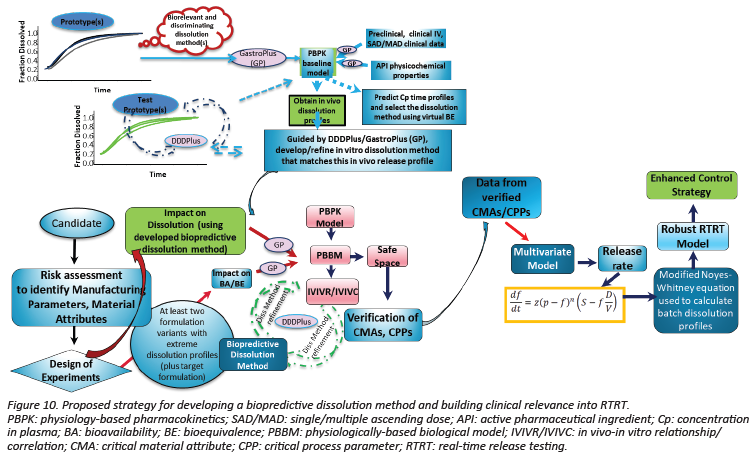
To date, few examples of dissolution models for real-time release testing (RTRT) have been approved for commercial drug products or published in literature. Thus, a structured approach has not been established by which a novice to the field could design, develop, validate, and implement an RTRT dissolution model. Moreover, with scant examples available, there has not been a body of work by which to learn of general regulatory expectations for such models. To address these gaps and to encourage conversation between regulatory and industrial experts on these topics, a virtual (web-based) workshop entitled “Predictive Dissolution Models for Real-Time Release Testing: Development and Implementation” was held November 11–12, 2021. This article summarizes key points from the podium presentations, panel discussions, and breakout sessions focusing on (1) the current best practices to establish predictive model specifications; (2) designing models to predict the “safe space” of a release test and creating models utilizing process analytical technology (PAT); and (3) exploring the strategy of compliant regulatory submissions, including model validation and post-approval lifecycle management. Industrial case studies were presented showcasing attempted approaches to and successful implementations of RTRT of dissolution for drug product manufacturing.
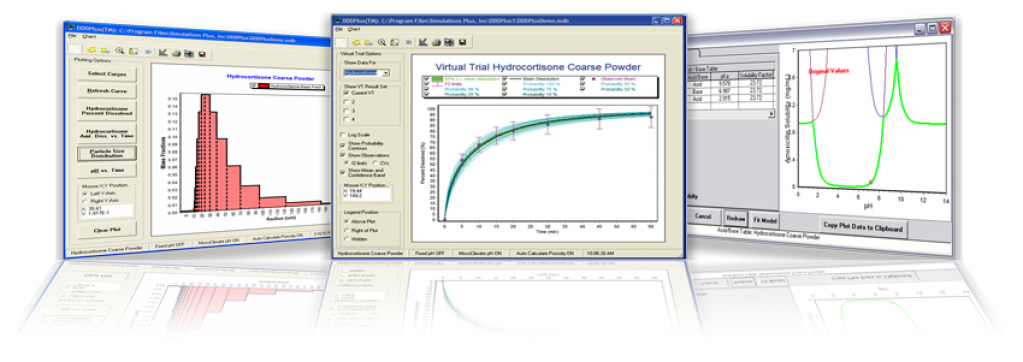
We have always been dedicated to carefully implement the best theories and develop novel approaches within the DDDPlus dissolution models. You enter limited physicochemical & manufacturing data, set up your dissolution method, and DDDPlus provides the rest:
- Simple, intuitive user interface
- Model optimization
- High-quality plots & figures for reporting purposes
- Excellent customer support
- Integration with our other tools, like ADMET Predictor® (QSAR) and GastroPlus® (PBPK & PBBM)
- The in vitro processes which are considered in the DDDPlus simulations, and the methods we’ve introduced to parameterize them, are too numerous to list here – instead, take a peek at the DDDPlus brochure for more details.
The DDDPlus dissolution modeling platform has been utilized by companies across various industries and departments since 2006. Some of the routine applications include:
- Assessing formulation strategies to achieve target dissolution profiles
- Assisting with dissolution method development
- Applying virtual ‘lot-to-lot’ variability effects to help establish dissolution specifications – remove the guesswork associated with the identification of dissolution variability and its impact on PK exposure
- Facilitating Quality by Design (QbD) implementation to guide product development
- Integrating with GastroPlus absorption, PBPK and PBBM models to optimize formulations and generate mechanistic in vitro-in vivo correlations (IVIVCs) – better extrapolation of dissolution inputs for PBPK & PBBM modeling… and more!
How do you spell relief?
Version 6 will continue providing the industry’s leading software for the in vitro dissolution experiment of pharmaceutical dosage forms! Model powders, capsules, both swellable and non-swellable and non-swellable polymer matrices, coated beads, delayed release, single or bilayer tablets and NEW controlled release and long acting injectable, dosage types.
NEW in DDDPlus version 6:
- New mechanistic Artificial Stomach-Duodenum model
- New in vitro dissolution apparatus models for improved IVIVE of precipitation kinetics with GastroPlus®:
- Biphasic dissolution
- Membrane dissolution
- New controlled release and Long Acting Injectable dosage form models
- and more!
Save time & money on your formulation design – request a software evaluation here.